Old Browser
Software and Informatics
BD Biosciences offers a suite of software and informatics support for flow cytometry analysis. From instrument-specific software to interfaces and powerful informatics, we support you every step of the way in your flow cytometry analysis.

Instrument Software
Our instrument software can support application set up, acquisition and analysis on our flow cytometers. Our IVD-cleared software, such as BD FACSCanto™ Clinical Software and the BD FACSuite™ Clinical Application support clinical applications while BD FACSChorus™ Software and BD FACSDiva™ Software when paired with research instruments, aid in research applications. All our software offer ease of use and help streamline your flow cytometry workflow.


Connectivity Software
Our connectivity software interface, such as the BD FACSLink™ Interface, helps connect your instruments to laboratory information systems (LIS). BD Assurity Linc™ Software connects compatible BD devices with BD technical support personnel so that they can rapidly troubleshoot your issues. The BD Remote Assist solution enables BD users to remotely connect to their BD devices from any computer.


FlowJo™ v10 Software
FlowJo™ v10 Software for research use features an intuitive interface, specialized analysis platforms and open-ended plugin architecture. It offers a rich analysis environment that is easy to use, versatile and extensible with plugins.


SeqGeq™ v1.6 Software
SeqGeq™ v1.6 Software is a device-agnostic desktop bioinformatics platform designed for the analysis of single-cell experiments and includes features such as V(D)J analysis, Seurat clustering, Monocle trajectory inference and much more.
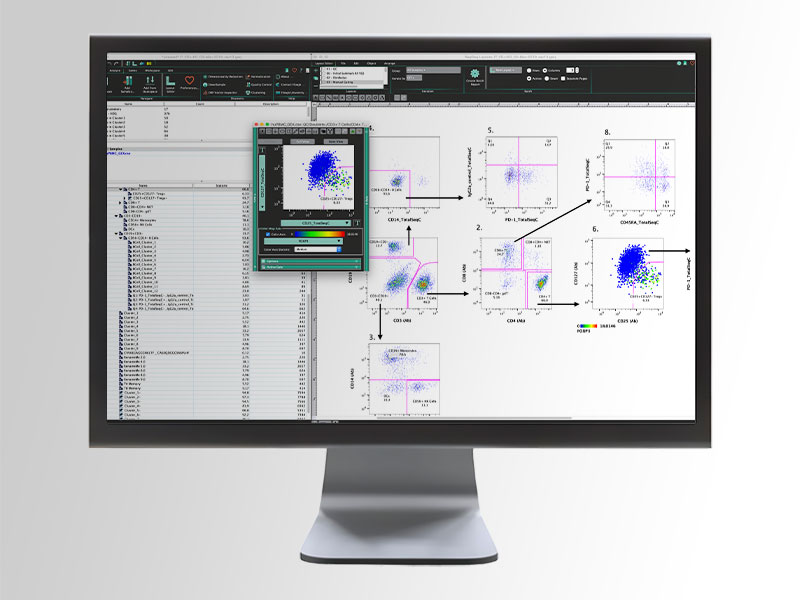

BD FACSDiva™ Software, when paired with research use instruments, BD FACSChorus™ Software, FlowJo™ Software and SeqGeq™ Software are for Research Use Only. Not for use in diagnostic or therapeutic procedures.
Report a Site Issue
This form is intended to help us improve our website experience. For other support, please visit our Contact Us page.