Old Browser
Overview
The BD Rhapsody™ Single-Cell Analysis System includes the BD Rhapsody™ Scanner and BD Rhapsody™ Express Single-Cell Analysis System. The BD Rhapsody™ Express System enables single-cell capture and barcoding of hundreds to thousands of single cells for analysis of genomic and proteomic information, using proprietary, gentle, robust microwell-based single-cell partitioning technology. The BD Rhapsody™ Scanner is designed to visualize all steps in the single-cell capture workflow and provide detailed analysis metrics at every step, enabling the user to make key decisions throughout the workflow.
Get more information from the BD Rhapsody™ System brochure.


FEATURES
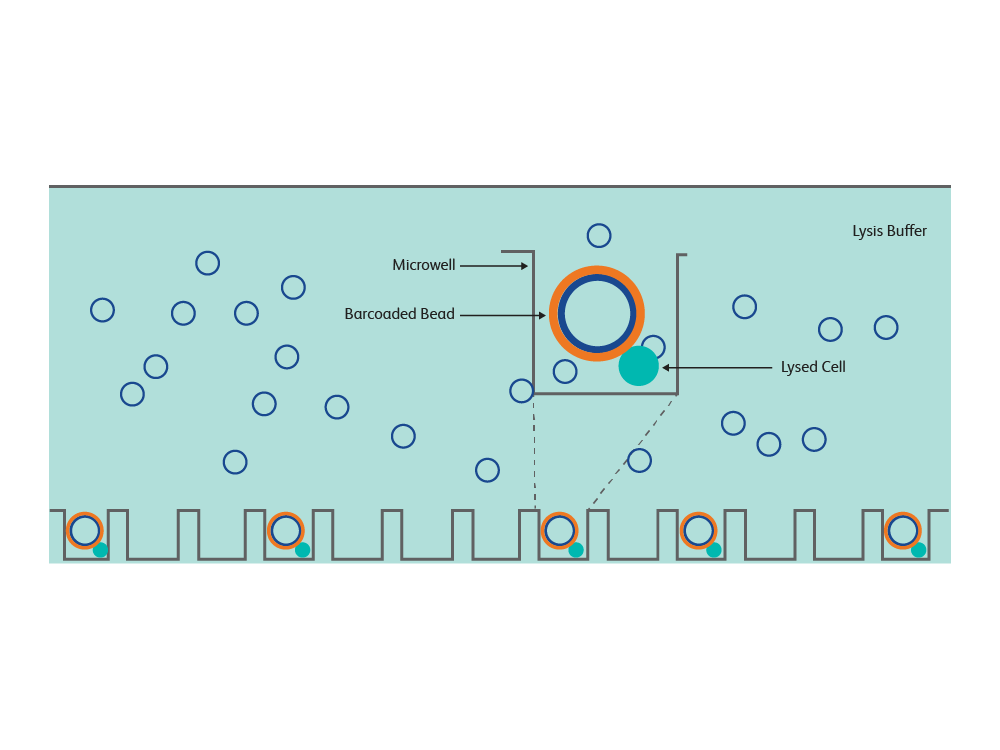

The BD Rhapsody™ Express Single-Cell Analysis System provides powerful microwell-based single-cell partitioning


- Minimal set of benchtop equipment, no fluidic pumps
- Allows gentle treatment of cells as they settle into microwells by gravity
- Each BD Rhapsody™ Cartridge contains a microwell array with >220,000 partitions
- The BD Rhapsody™ Cartridge has a volume ~600 µL; dilute cell suspensions can be loaded directly into the cartridge without needing to concentrate them into very small volumes
- No microfluidic channels are involved so clogging of channels with cells is less of a concern
- The microwells are specially treated to prevent cells from adhering to surfaces and extensive QC is performed
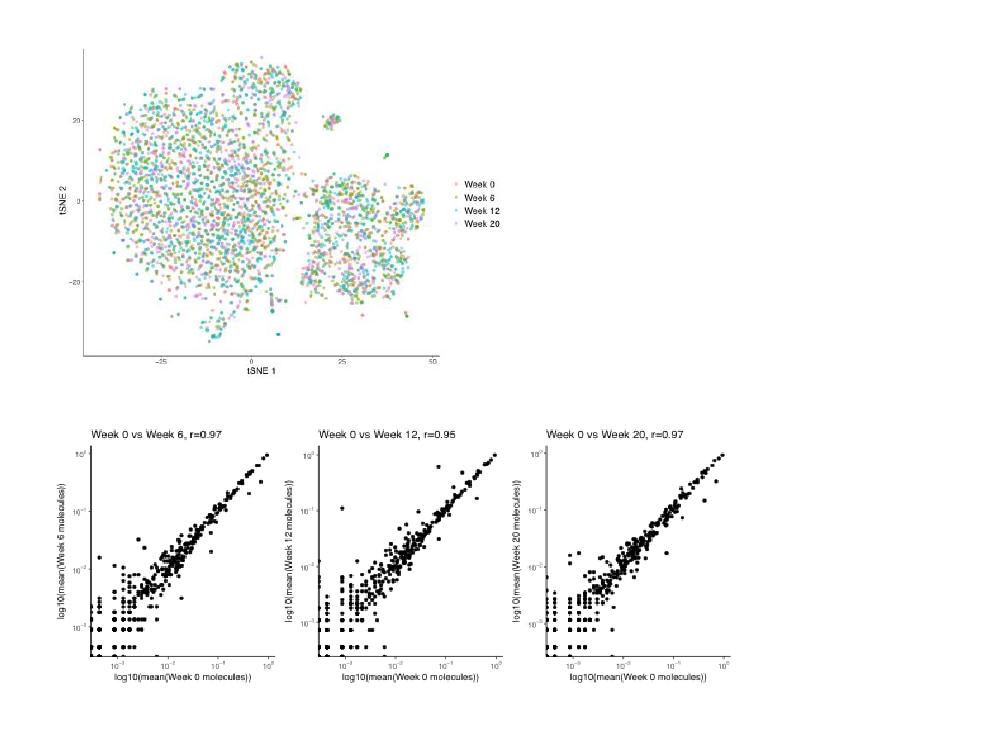

Stable magnetic barcoded beads used with the BD Rhapsody™ System can be retained for later experimentation allowing for flexibility with experimental design


- BD Rhapsody™ Beads remain intact throughout the workflow
- Beads with cDNA molecules can be stored for several months
- Beads can be subsampled for creation of multiple sequencing libraries
- Archived beads can be pooled and sequenced together for operational efficiency
- Archived beads can be shared easily with research partners
- Archived, subsampled beads can help standardize performance and develop SOPs
Right: Image showing high correlation in gene expression and minimal batch effects seen in data obtained from beads stored up to 12 weeks
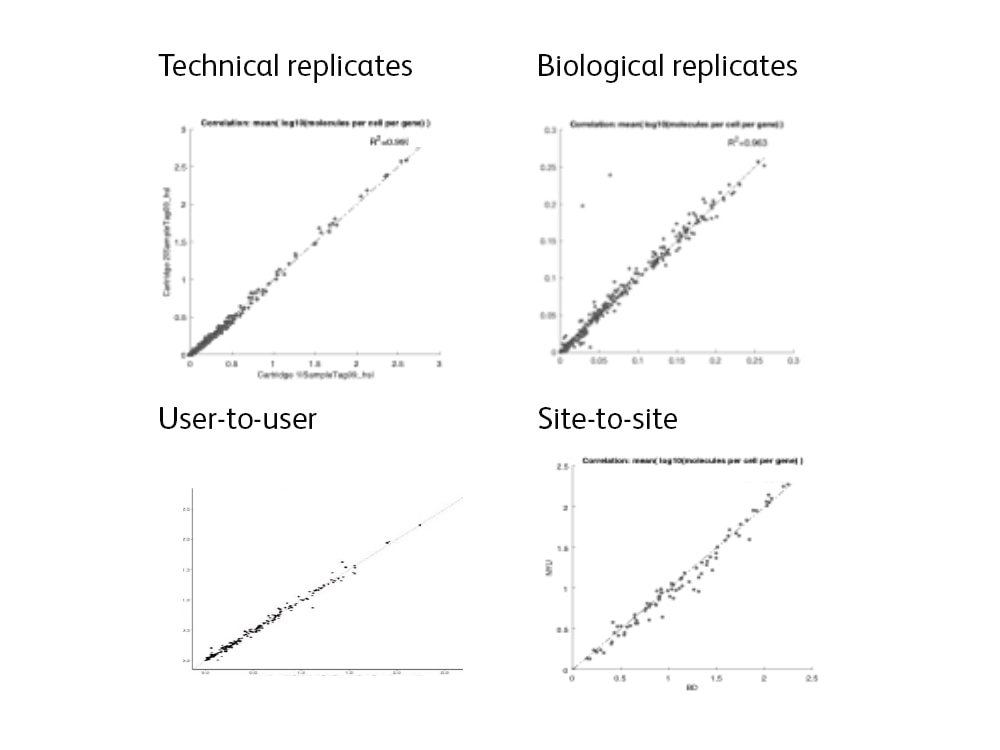

The BD Rhapsody™ Single-Cell Analysis System is the ideal choice for translational, multi-center, multi-cohort studies


Reliable, reproducible results from gentle, gravity-based single-cell partitioning without microfluidics
- Minimal batch effects with technical replicates
- Minimal batch effects with biological replicates
- Minimal site-to-site variability
- Minimal user-to-user variability
- Minimum sample bias
- Ability to integrate large data sets without correction for batch effects
Right: Image showing high correlation in gene expression seen between technical and biological replicates and between users and sites.
The BD Rhapsody™ Scanner provides the ability to visualize single-cell capture and provides key quality metrics at every step
The BD Rhapsody™ Scanner:
- Enables visual inspection of the cartridge and of each microwell through an optically clear window on the BD Rhapsody™ Cartridge
- Provides quality control measures such as cell capture rate and cell multiplet rates by direct imaging of the cartridge to analyze quality of the cell preparation. Quality control metrics provide an indication if errors have occurred during the cartridge workflow allowing you to decide whether to proceed to sequencing
- Can be useful for troubleshooting and provide an estimate of the number of cells expected to be retrieved by sequencing
Analysis
| Number of wells with viable cells at cell load 9118 |
| Cell multiplet rate at cell load 2.4% |
| Number of wells with viable cells and a bead 7999 |
| Cell multiplet rate 2.0% |
| Bead loading efficiency PASS |
| Cell retention rate PASS |
| Bead retrieval efficiency PASS |
| Excess bead rate PASS |
PERFORMANCE
The BD Rhapsody™ Single-Cell Analysis System provides reliable and reproducible results making it an ideal system for translational studies
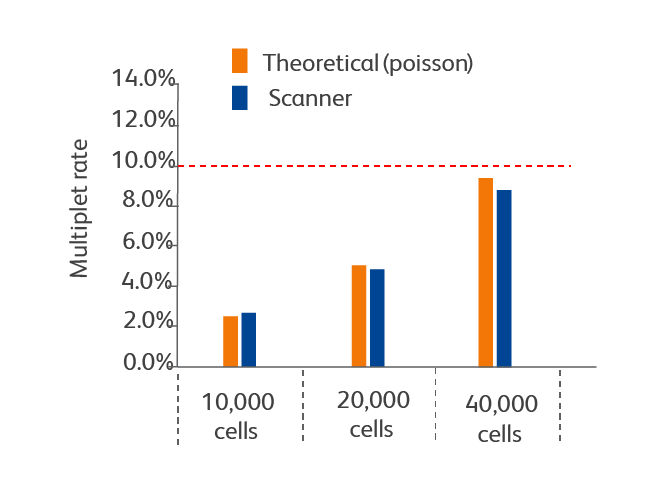
The system offers superior cell capture rates and low multiplet rates with a variety of cell types as compared to other systems
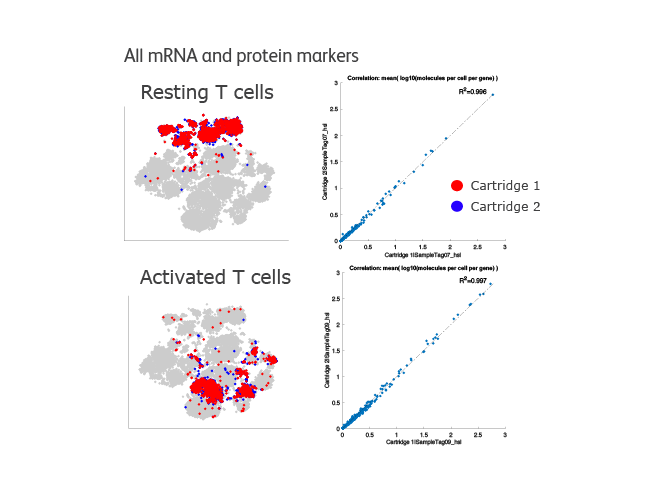
Strong correlation of all mRNA and protein markers identifies similar T-cell populations between cartridges
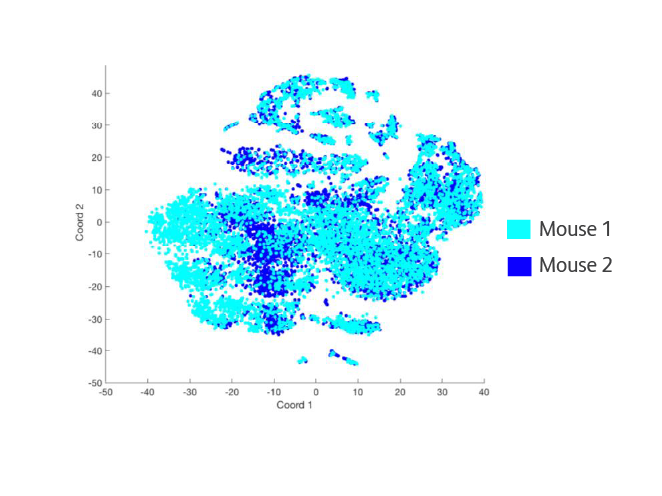
Equivalent data from biological replicates
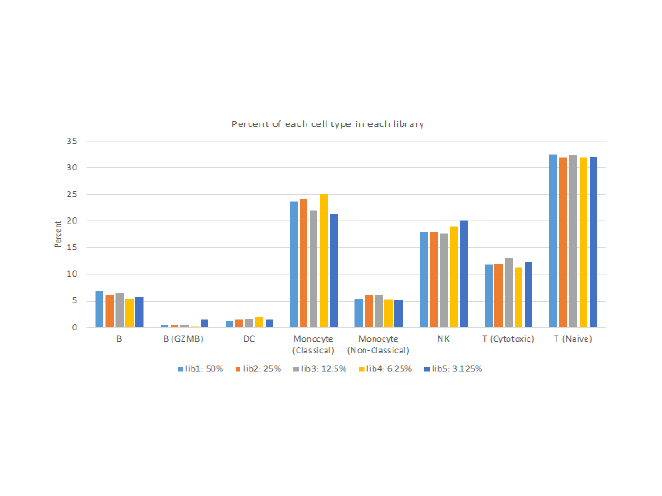
Equivalent data from subsampled beads
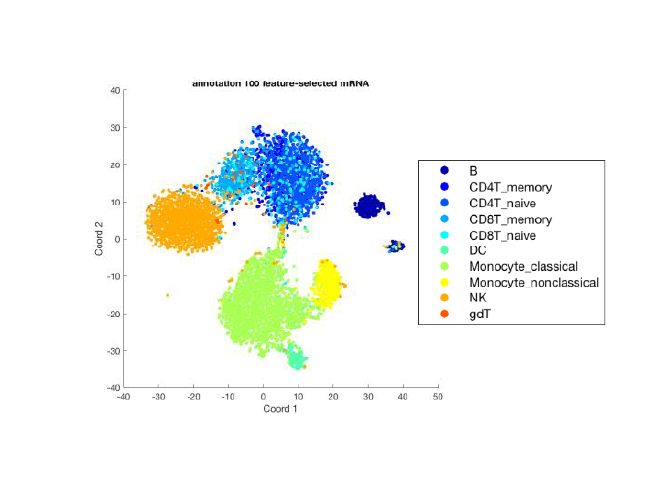
Similar proportions of each cell type are recovered from WTA and targeted assays performed on subsampled beads.
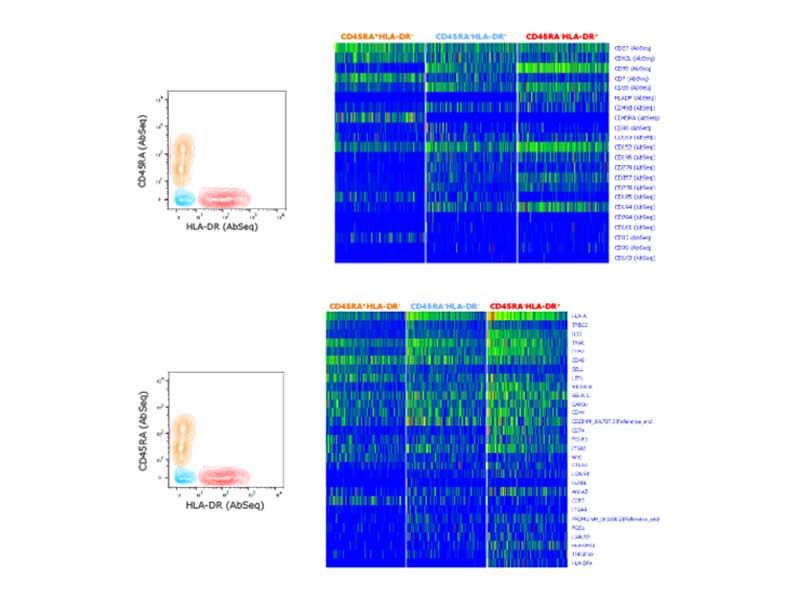
The BD Rhapsody™ Single-Cell Analysis System enables simultaneous measurement of surface proteins and mRNA expression, facilitating the identification of distinct subsets of regulatory T cells
Differential gene expression and differential protein expression analysis were performed on three main Treg subsets identified based on manual gating and bivariate analysis of CD45RA and HLA-DR sorted Treg samples. Differential gene expression analysis was performed using the BD Rhapsody™ Targeted Human Immune Response Panel (399 genes). Upregulated genes (>1.25X) are shown in the heat map above. Differential protein expression of 22 proteins was performed simultaneously using BD® AbSeq Oligo-Conjugated Antibodies.

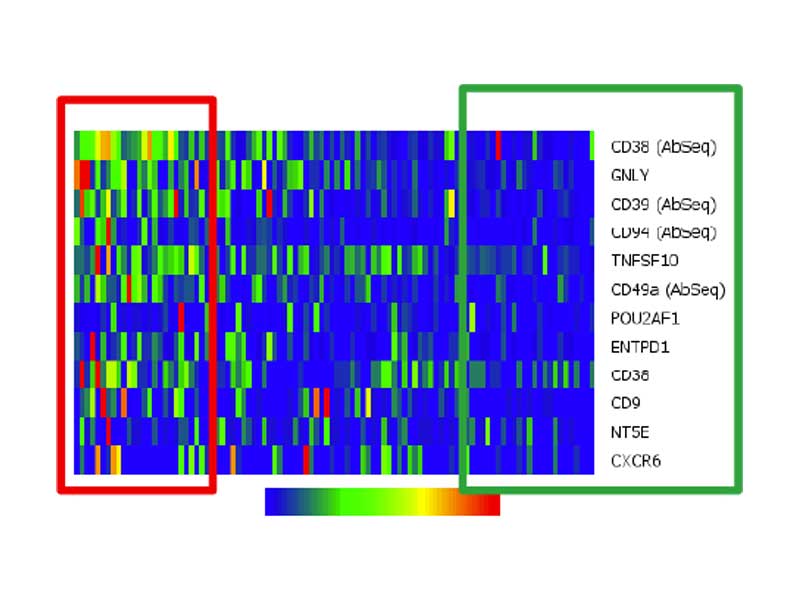
The BD Rhapsody™ Single-Cell Analysis System can be used for deep characterization of exhausted T cells
Expression of 12 proteins and genes that are significantly upregulated at day 14 of chronic simulation of CD8+ T cells. Two populations (red and green boxes) emerged based on the expression level of markers. Co-expression patterns were discerned for CD38, CD39 and CD94.

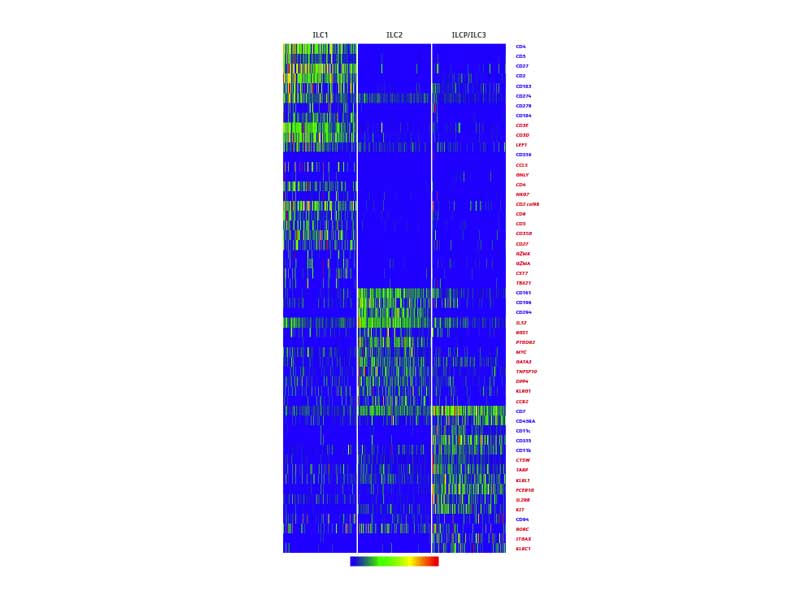
The BD Rhapsody™ Single-Cell Analysis System enables resolution of innate lymphoid cell heterogeneity
Differential protein and gene expression at the single-cell level.
Heterogeneity was observed in major ILC subsets.

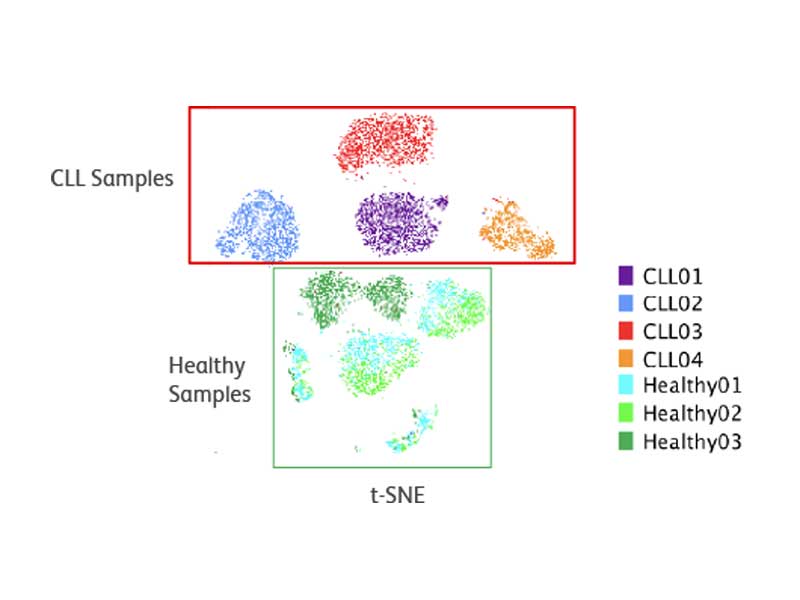
The BD Rhapsody™ Single-Cell Analysis System enables identification of heterogeneity within CLL samples
Clusters are well defined using simultaneous mRNA and protein analysis. Samples from healthy donors cluster similarly. CLL samples cluster separately and uniquely.

-
Brochures
-
User Guides
-
Data Sheets
-
A Comprehensive Characterization of Regulatory T Cells Using the BD Rhapsody™ Single-Cell Analysis System
-
Assessing Obesity-Induced Inflammation in Mice through Single-Cell Multiomic Analysis
-
Exploring Tumor Heterogeneity of Chronic Lymphocytic Leukemia Using Single-Cell Multiomics
-
Resolving Human Circulating Innate Lymphoid Heterogeneity Using Advanced Single-Cell Multiomic Analysis
-
Single-Cell Multiomic Analysis of T Cell Exhaustion In Vitro
-
Streamlined Transfer of High-Parameter Flow Cytometry Panel from Cell Analyzer to Cell Sorter Prior to In-Depth Single-Cell Multiomic Analysis of Murine Lymphocyte Subpopulations
-
Leveraging the Power of High-Parameter Cell Sorting and Single-Cell Multiomics to Profile Intratumoral Immune Cells in a Model of B-Cell Lymphoma
-
Presentations
-
CLL Phenotyping with BD Rhapsody™ Targeted Panels and BD® AbSeq Assays
-
Single-Cell Multiomic Analysis for Deep Characterization of Exhausted T Cells
-
Single-Cell Multiomic Analysis of Immune Cells in Obese Mice
-
One Solution for Comprehensive Treg Characterization Via Single-Cell Multiomic Analysis
-
Single-Cell Multiomic Analysis for the Resolution of Innate Lymphoid Cell Heterogeneity
-
Protocols
Playlist contains:
- Counting Cells with the BD Rhapsody™ Single-Cell Analysis System
- BD Rhapsody™ Single-Cell Analysis System Post PCR1 Purification
- Capturing Single Cells with the BD Rhapsody™ Express Single-Cell Analysis System
- Performing Quality Control on the Final Sequencing Libraries
- Single-Cell Labeling with BD® AbSeq Ab-Oligos and the BD® Single-Cell Multiplexing Kit
- BD Rhapsody™ Bioinformatics Pipeline
- One Workflow: Cell Enrichment to Single Cell Transcriptome and Proteome Analysis

For Research Use Only. Not for use in diagnostic or therapeutic procedures.
Results shown are not guaranteed and may not be representative.
Dynabeads is a trademark of Thermo Fisher Scientific.
Report a Site Issue
This form is intended to help us improve our website experience. For other support, please visit our Contact Us page.